Figures of the Article
-
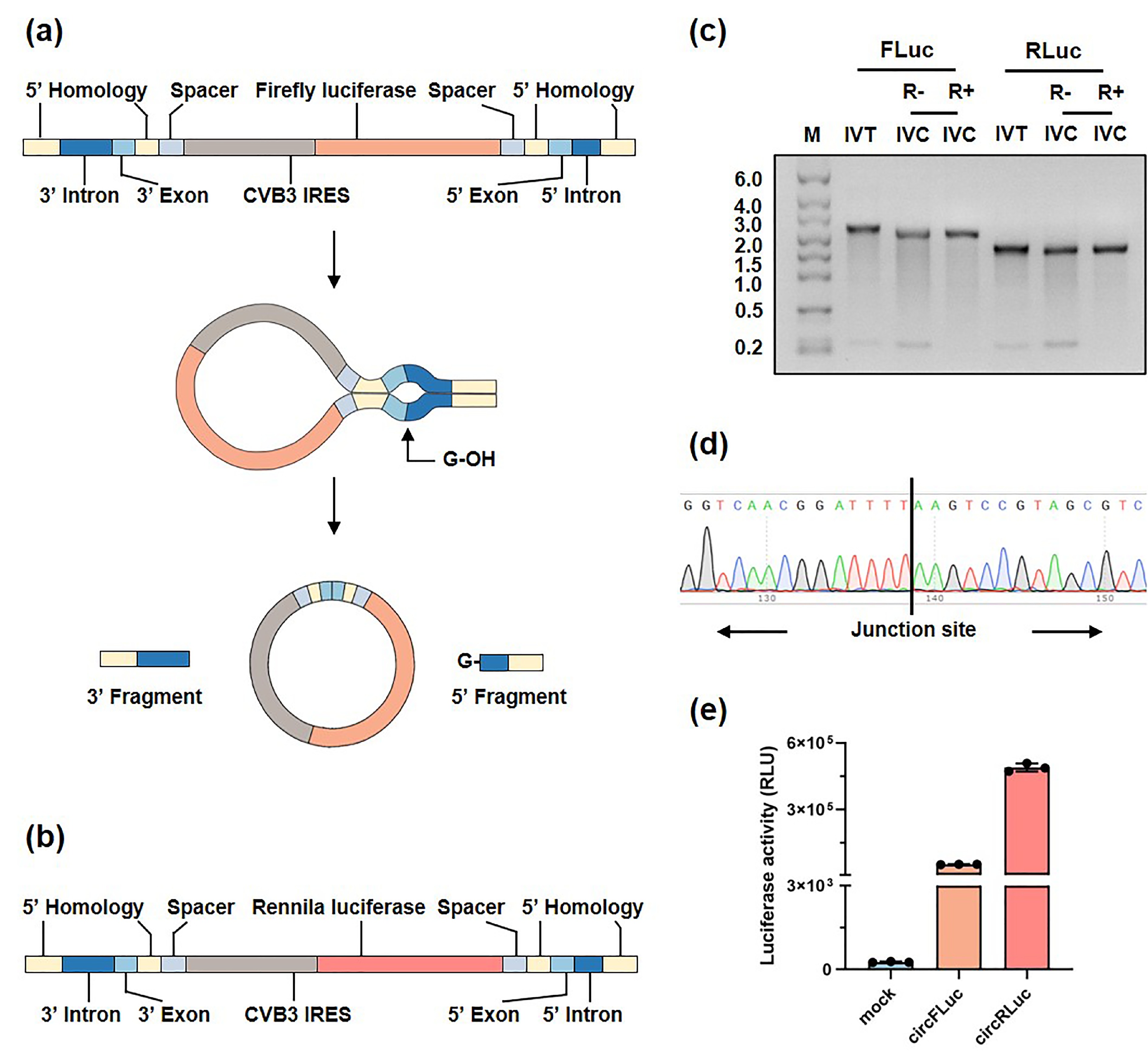
![]() Protein expression initiated by circRNAs. (a, b) Schematic representation of the elements required for circFLuc (a) and circRLuc (b) production through the PIE system. (c) Agarose gel showing linear RNA after in vitro transcription (IVT) and untreated and treated in vitro cyclization (IVC) RNA products with RNase R. (d) Sanger sequencing chromatogram for the junction site of the reverse-transcribed circFLuc and circRLuc samples. (e) Luciferase activity (RLU) of circFLuc and circRLuc in HEK293T cells. Statistical analysis of luciferase activity was performed at 24 h posttransfection. FLuc represents firefly luciferase. RLuc represents Renilla luciferase. RLU, relative light unit.
Protein expression initiated by circRNAs. (a, b) Schematic representation of the elements required for circFLuc (a) and circRLuc (b) production through the PIE system. (c) Agarose gel showing linear RNA after in vitro transcription (IVT) and untreated and treated in vitro cyclization (IVC) RNA products with RNase R. (d) Sanger sequencing chromatogram for the junction site of the reverse-transcribed circFLuc and circRLuc samples. (e) Luciferase activity (RLU) of circFLuc and circRLuc in HEK293T cells. Statistical analysis of luciferase activity was performed at 24 h posttransfection. FLuc represents firefly luciferase. RLuc represents Renilla luciferase. RLU, relative light unit.
-
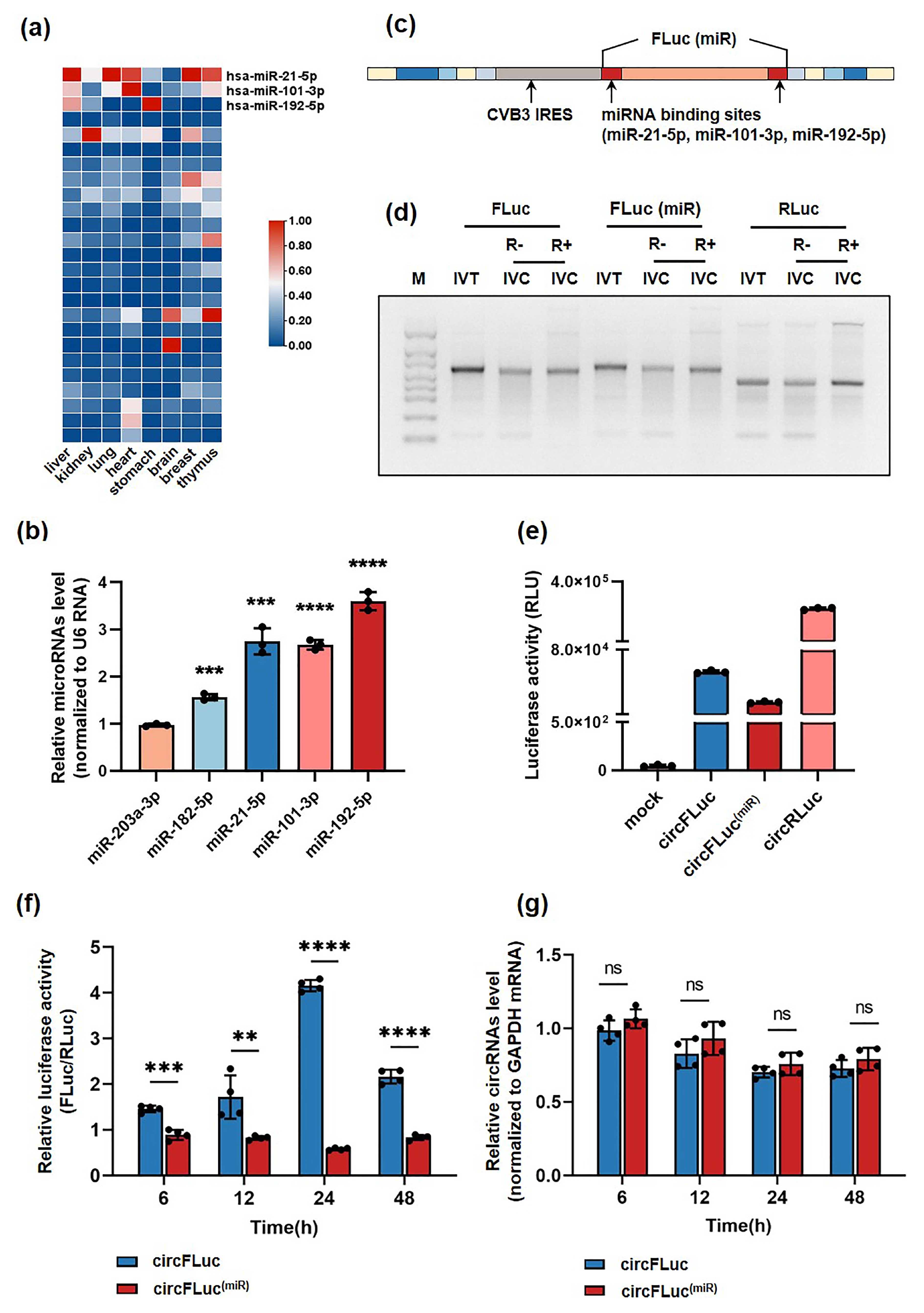
![]() Translation efficacy of circFLuc(miR) and circFLuc. (a) Heatmap displaying the expression profile of miRNAs in human healthy tissues. Data were normalized using the Z-score method. (b) Relative expression levels of microRNAs in the database were determined by RT‒qPCR, with U6 RNA serving as an internal control (n = 3). (c) Schematic diagram of the circularized sequences after the addition of miRNA binding sites (miR-21-5p, miR-101-3p, miR-192-5p). (d) Agarose gel showing linear RNA after in vitro transcription (IVT) and untreated and treated in vitro cyclization (IVC) RNA products with RNase R. (e) Luciferase activity (RLU) of circFLuc(miR), circFLuc, and circRLuc in HEK293T cells. Activity was measured at 24 h posttransfection. (f) Relative luciferase activity of 293T cells transfected with circFLuc(miR) and circFLuc, with cotransfected circRLuc serving as an internal control, and relative firefly luciferase activity normalized to RLuc (n = 4). (g) Relative circRNA levels in HEK293T cells transfected with circFLuc(miR) and circFLuc, normalized to GAPDH mRNA and circRLuc (n = 4). Statistical analyses were performed at different time points. RLU, relative light unit. ns represents no significant difference, *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Translation efficacy of circFLuc(miR) and circFLuc. (a) Heatmap displaying the expression profile of miRNAs in human healthy tissues. Data were normalized using the Z-score method. (b) Relative expression levels of microRNAs in the database were determined by RT‒qPCR, with U6 RNA serving as an internal control (n = 3). (c) Schematic diagram of the circularized sequences after the addition of miRNA binding sites (miR-21-5p, miR-101-3p, miR-192-5p). (d) Agarose gel showing linear RNA after in vitro transcription (IVT) and untreated and treated in vitro cyclization (IVC) RNA products with RNase R. (e) Luciferase activity (RLU) of circFLuc(miR), circFLuc, and circRLuc in HEK293T cells. Activity was measured at 24 h posttransfection. (f) Relative luciferase activity of 293T cells transfected with circFLuc(miR) and circFLuc, with cotransfected circRLuc serving as an internal control, and relative firefly luciferase activity normalized to RLuc (n = 4). (g) Relative circRNA levels in HEK293T cells transfected with circFLuc(miR) and circFLuc, normalized to GAPDH mRNA and circRLuc (n = 4). Statistical analyses were performed at different time points. RLU, relative light unit. ns represents no significant difference, *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
-
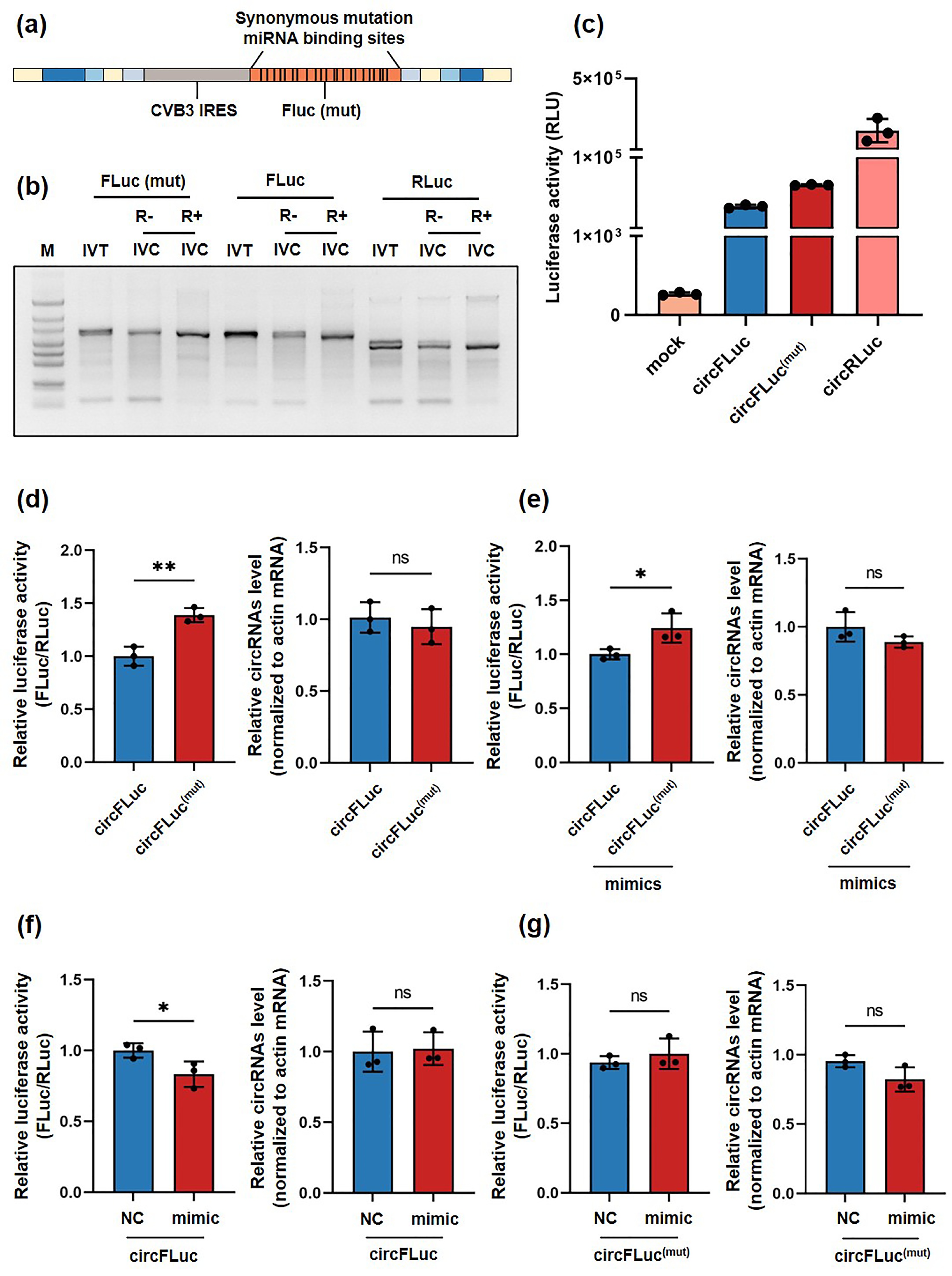
![]() Translation efficacy of circFLuc(mut) and circFLuc. (a) Schematic illustration of circularized sequences with synonymous mutations introduced in miRNA binding sites. (b) Agarose gel showing linear RNA after in vitro transcription (IVT) and untreated and treated in vitro cyclization (IVC) RNA products with RNase R. (c) Luciferase activity (RLU) of circFLuc(mut), circFLuc and circRLuc in HEK293T cells at 24 h posttransfection. (d–g) Left panel shows the relative luciferase activity of HEK293T cells transfected with circFLuc(mut) or circFLuc (n = 3). CircRLuc was used as an internal control for cotransfection, while miR-129-5p and miR-362-5p were overexpressed through mimics (mimic). The nonoverexpressing group was transfected with NC-mimics as a control (NC). The relative luciferase activity was standardized using RLuc activity. The right panel shows the relative expression level of circRNAs in transfected cells (n = 3) normalized to actin mRNA and circRLuc. Statistical analysis was performed at 24 h posttransfection. RLU, relative light unit. ns, not significant, *p < 0.05, ** p < 0.01.
Translation efficacy of circFLuc(mut) and circFLuc. (a) Schematic illustration of circularized sequences with synonymous mutations introduced in miRNA binding sites. (b) Agarose gel showing linear RNA after in vitro transcription (IVT) and untreated and treated in vitro cyclization (IVC) RNA products with RNase R. (c) Luciferase activity (RLU) of circFLuc(mut), circFLuc and circRLuc in HEK293T cells at 24 h posttransfection. (d–g) Left panel shows the relative luciferase activity of HEK293T cells transfected with circFLuc(mut) or circFLuc (n = 3). CircRLuc was used as an internal control for cotransfection, while miR-129-5p and miR-362-5p were overexpressed through mimics (mimic). The nonoverexpressing group was transfected with NC-mimics as a control (NC). The relative luciferase activity was standardized using RLuc activity. The right panel shows the relative expression level of circRNAs in transfected cells (n = 3) normalized to actin mRNA and circRLuc. Statistical analysis was performed at 24 h posttransfection. RLU, relative light unit. ns, not significant, *p < 0.05, ** p < 0.01.
-
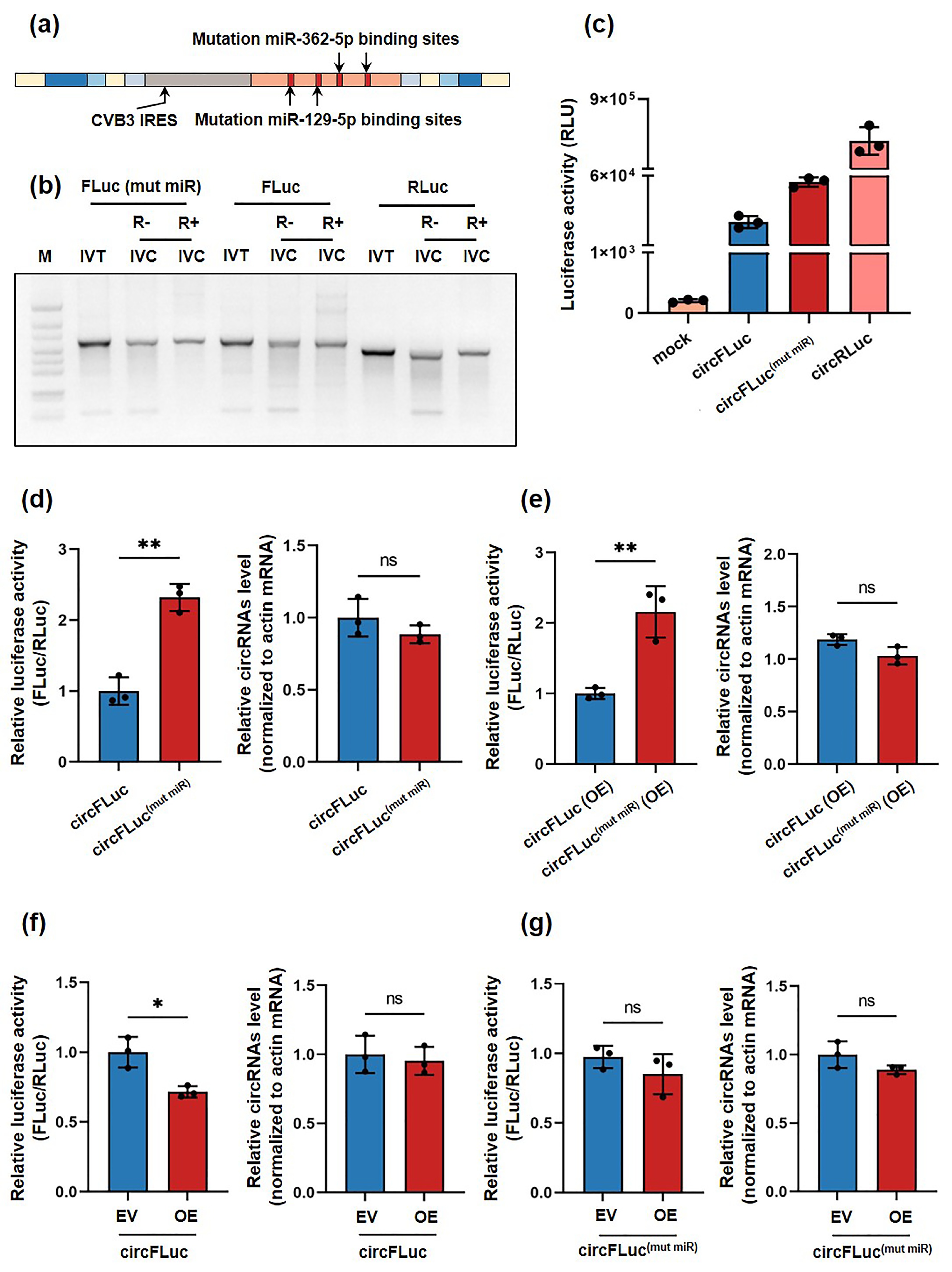
![]() Translation efficacy of circFLuc(mut miR) and circFLuc. (a) Schematic diagram of the circularized sequence with synonymous mutations in the miR-129-5p and miR-362-5p binding sites. (b) Agarose gel showing linear RNA after in vitro transcription (IVT) and untreated and treated in vitro cyclization (IVC) RNA products with RNase R. (c) Luciferase activity (RLU) of circFLuc(mut miR), circFLuc, and circRLuc in HEK293T cells, with detection of luciferase activity at 24 h posttransfection. (d–g) The left panel shows the relative luciferase activity of HEK293T cells (n = 3) transfected with circFLuc(mut miR) or circFLuc. CircRLuc was used as an internal control for cotransfection, while miR-129-5p and miR-362-5p were overexpressed through plasmids (OE). The nonoverexpressing group was transfected with an empty plasmid backbone as a control (EV). The relative luciferase activity was standardized using RLuc activity. The right panel shows that the relative circRNA level was standardized using actin mRNA and circRLuc. Statistical analysis was performed at 24 h posttransfection. RLU, relative light unit. ns, no significance, *p < 0.05, ** p < 0.01.
Translation efficacy of circFLuc(mut miR) and circFLuc. (a) Schematic diagram of the circularized sequence with synonymous mutations in the miR-129-5p and miR-362-5p binding sites. (b) Agarose gel showing linear RNA after in vitro transcription (IVT) and untreated and treated in vitro cyclization (IVC) RNA products with RNase R. (c) Luciferase activity (RLU) of circFLuc(mut miR), circFLuc, and circRLuc in HEK293T cells, with detection of luciferase activity at 24 h posttransfection. (d–g) The left panel shows the relative luciferase activity of HEK293T cells (n = 3) transfected with circFLuc(mut miR) or circFLuc. CircRLuc was used as an internal control for cotransfection, while miR-129-5p and miR-362-5p were overexpressed through plasmids (OE). The nonoverexpressing group was transfected with an empty plasmid backbone as a control (EV). The relative luciferase activity was standardized using RLuc activity. The right panel shows that the relative circRNA level was standardized using actin mRNA and circRLuc. Statistical analysis was performed at 24 h posttransfection. RLU, relative light unit. ns, no significance, *p < 0.05, ** p < 0.01.
-
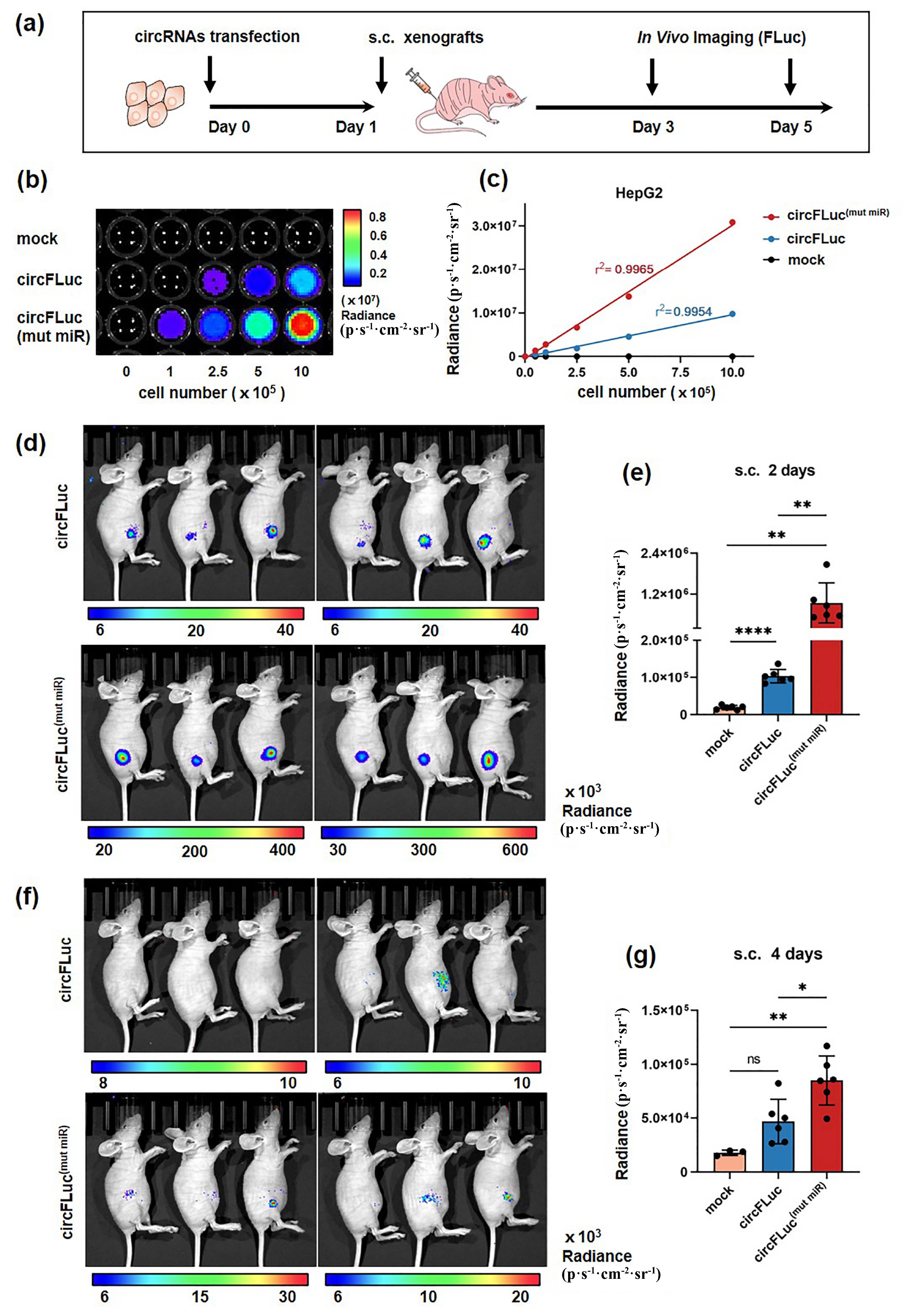
![]() In vivo translation efficiency of circFLuc(mut miR) and circFLuc. (a) Schematic diagram of the in vivo experiment in nude mice. (b) Bioluminescence imaging of HepG2 cells (top), HepG2-circFLuc cells (middle), and HepG2-circFLuc(mut miR) cells (bottom) was performed. Luciferase activity was detected 24 h after transfection of different cell numbers. Images show color-coded maps of photon flux superimposed on black-and-white photographs of the assay plates. (c) The statistical results of (b) show that the bioluminescence values of circFLuc(mut miR) and circFLuc are linearly correlated with the number of cells, with r2 values of 0.9965 and 0.9954, respectively. Luminescence values in the mock group did not change with increasing cell number. (d) In vivo bioluminescence image of mice subcutaneously injected with HepG2-circFLuc (top) and HepG2-circFLuc(mut miR) (bottom) and measured for luciferase activity 2 d post-injection (n = 6 per group, 106 cells per mouse). (e) Statistical analysis of the in vivo luminescence values from (d). (f) In vivo bioluminescence image of mice subcutaneously injected with HepG2-circFLuc (top) and HepG2-circFLuc(mut miR) (bottom) and measured for luciferase expression 4 d post-injection (n = 6 per group, 106 cells per mouse). (g) Statistical analysis of the in vivo luminescence values from (f). ns, no significance, *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
In vivo translation efficiency of circFLuc(mut miR) and circFLuc. (a) Schematic diagram of the in vivo experiment in nude mice. (b) Bioluminescence imaging of HepG2 cells (top), HepG2-circFLuc cells (middle), and HepG2-circFLuc(mut miR) cells (bottom) was performed. Luciferase activity was detected 24 h after transfection of different cell numbers. Images show color-coded maps of photon flux superimposed on black-and-white photographs of the assay plates. (c) The statistical results of (b) show that the bioluminescence values of circFLuc(mut miR) and circFLuc are linearly correlated with the number of cells, with r2 values of 0.9965 and 0.9954, respectively. Luminescence values in the mock group did not change with increasing cell number. (d) In vivo bioluminescence image of mice subcutaneously injected with HepG2-circFLuc (top) and HepG2-circFLuc(mut miR) (bottom) and measured for luciferase activity 2 d post-injection (n = 6 per group, 106 cells per mouse). (e) Statistical analysis of the in vivo luminescence values from (d). (f) In vivo bioluminescence image of mice subcutaneously injected with HepG2-circFLuc (top) and HepG2-circFLuc(mut miR) (bottom) and measured for luciferase expression 4 d post-injection (n = 6 per group, 106 cells per mouse). (g) Statistical analysis of the in vivo luminescence values from (f). ns, no significance, *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
-
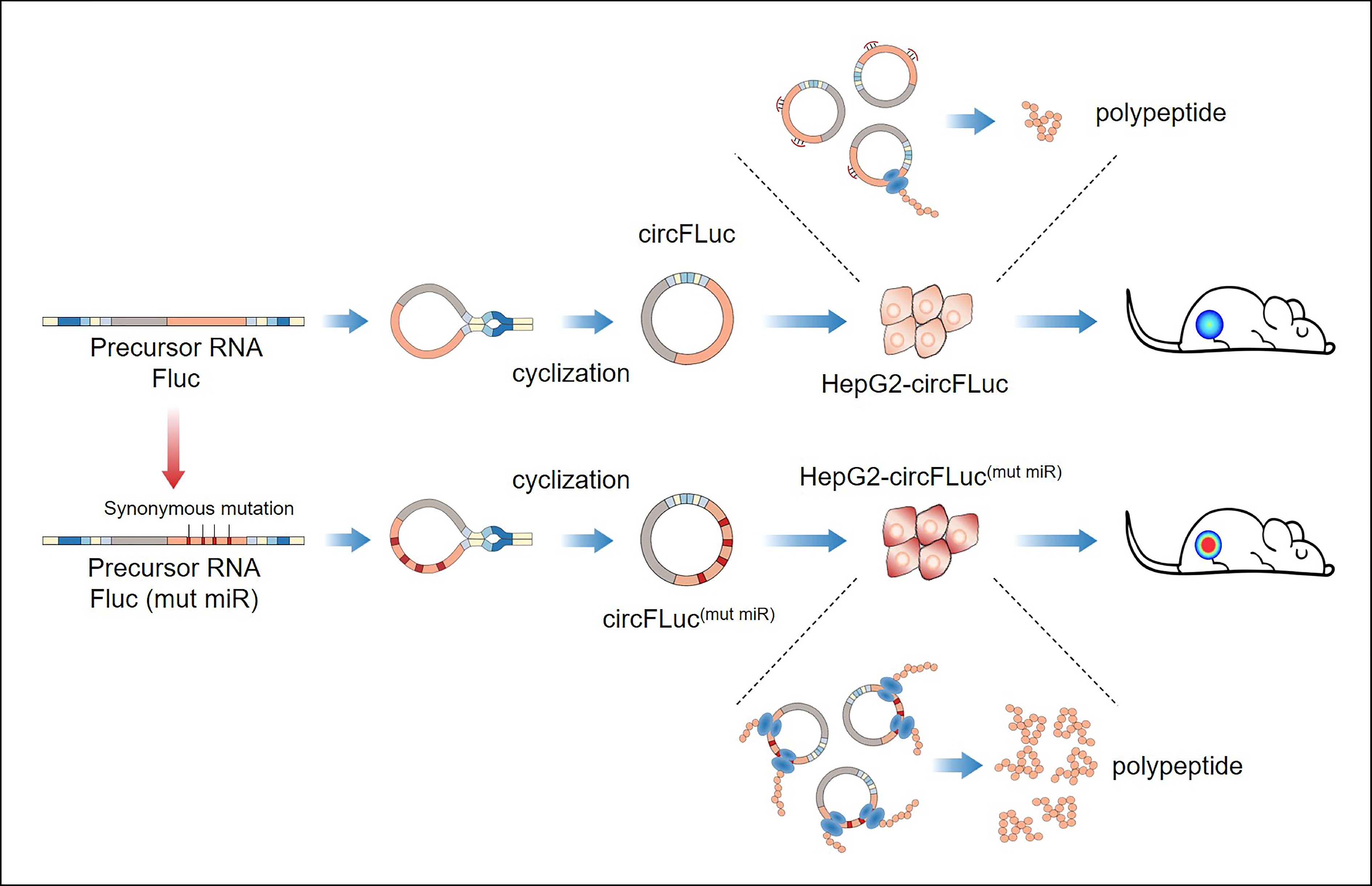
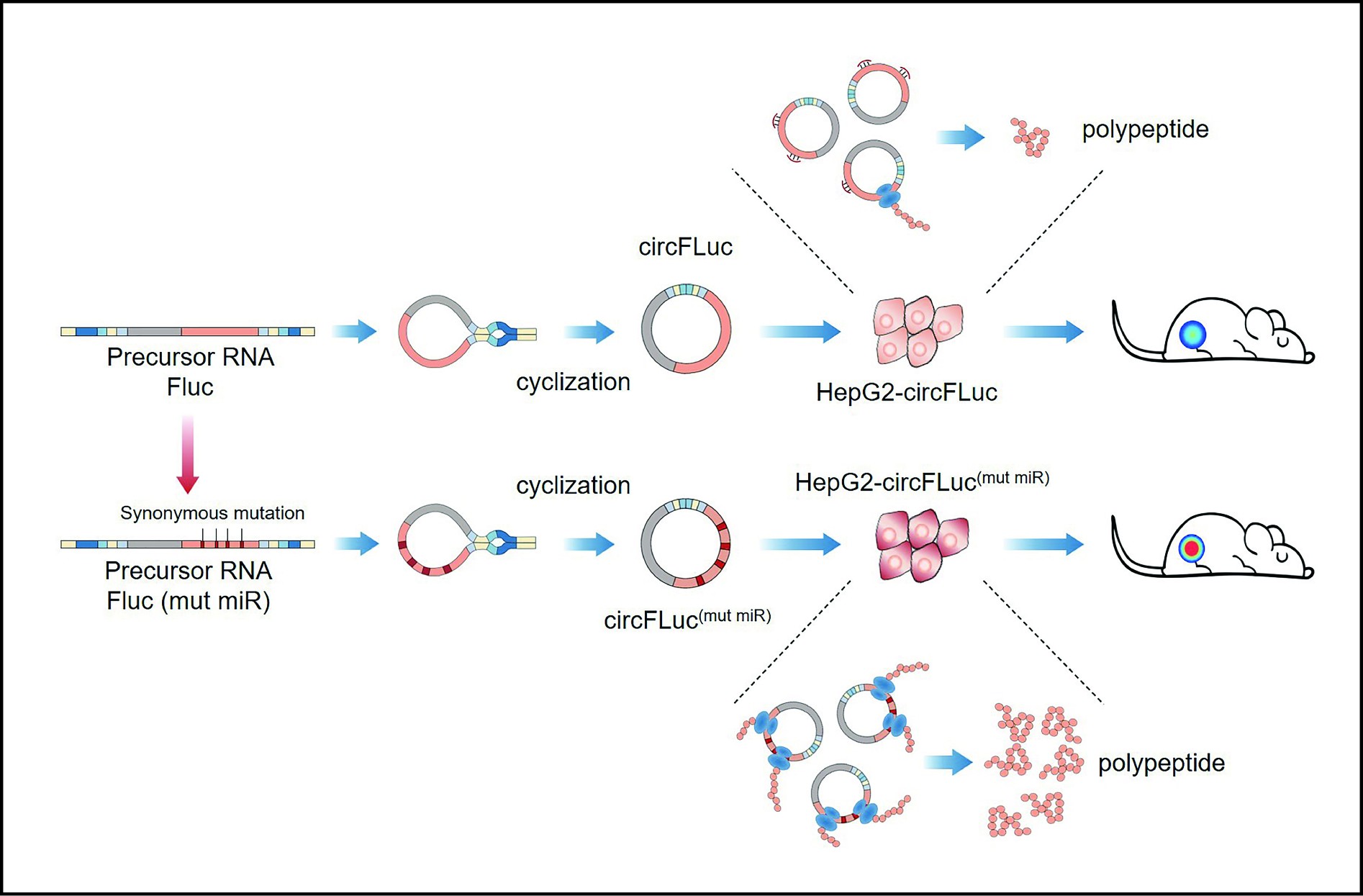
![]() A simplified summary of this study, in vitro synthesis of circRNA and related changes in miRNA binding sites and their roles in vitro and in vivo.
A simplified summary of this study, in vitro synthesis of circRNA and related changes in miRNA binding sites and their roles in vitro and in vivo.
Related articles
-
2021, 51(10): 747-752. DOI: 10.52396/JUST-2021-0032
-
2020, 50(7): 1013. DOI: 10.3969/j.issn.0253-2778.2020.07.019
-
2020, 50(4): 457-466. DOI: 10.3969/j.issn.0253-2778.2020.04.010
-
2018, 48(9): 755-761. DOI: 10.3969/j.issn.0253-2778.2018.09.010
-
2018, 48(9): 723-729. DOI: 10.3969/j.issn.0253-2778.2018.09.006
-
2017, 47(4): 283-289. DOI: 10.3969/j.issn.0253-2778.2017.04.001
-
2016, 46(3): 222-230. DOI: 10.3969/j.issn.0253-2778.2016.03.007
-
2016, 46(1): 66-75. DOI: 10.3969/j.issn.0253-2778.2016.01.009
-
2015, 45(1): 56-60. DOI: 10.3969/j.issn.0253-2778.2015.01.009
-
2013, 43(11): 933-940. DOI: 10.3969/j.issn.0253-2778.2013.11.009



 Download:
Download: