ISSN 0253-2778
CN 34-1054/N
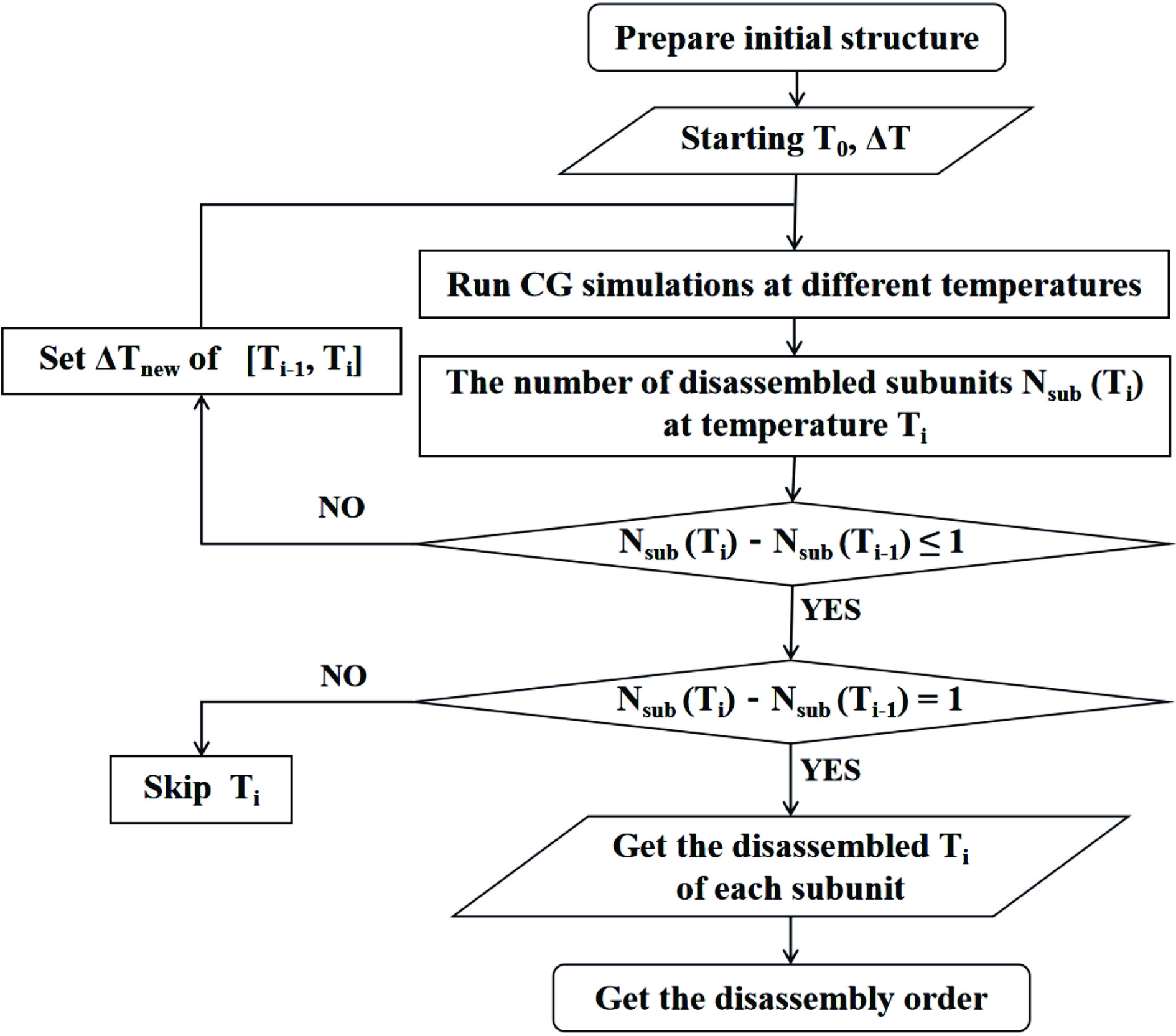
The assembly of a protein complex is very important for its biological function, which can be investigated by determining the order of assembly/disassembly of its protein subunits. Although static structures of many protein complexes are available in the protein data bank, their assembly/disassembly orders of subunits are largely unknown. In addition to experimental techniques for studying subcomplexes in the assembly/disassembly of a protein complex, computational methods can be used to predict the assembly/disassembly order. Since sampling is a nontrivial issue in simulating the assembly/disassembly process, coarse-grained simulations are more efficient than atomic simulations are. In this work, we developed computational protocols for predicting the assembly/disassembly orders of protein complexes via coarse-grained simulations. The protocols were illustrated via two protein complexes, and the predicted assembly/disassembly orders were consistent with the available experimental data.

The assembly and disassembly orders of the Arp2/3 complex were predicted via coarse-grained simulations.
Figure 8. Stability of subcomplexes during the assembly of the Arp2/3 complex. The mean and standard deviation of the RMSD value of each subcomplex are shown from (a) the second to (d) the fifth iteration. For each subcomplex, the RMSD was calculated by taking the average of three independent CG simulations.
| [1] |
Sali A, Glaeser R, Earnest T, et al. From words to literature in structural proteomics. Nature, 2003, 422 (6928): 216–225. DOI: 10.1038/nature01513
|
| [2] |
Guaita M, Watters S C, Loerch S. Recent advances and current trends in cryo-electron microscopy. Current Opinion in Structural Biology, 2022, 77: 102484. DOI: 10.1016/j.sbi.2022.102484
|
| [3] |
Shi Y. A glimpse of structural biology through X-ray crystallography. Cell, 2014, 159 (5): 995–1014. DOI: 10.1016/j.cell.2014.10.051
|
| [4] |
Gao M, Nakajima An D, Parks J M, et al. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nature Communications, 2022, 13: 1744. DOI: 10.1038/s41467-022-29394-2
|
| [5] |
Marsh J A, Teichmann S A. Structure, dynamics, assembly, and evolution of protein complexes. Annual Review of Biochemistry, 2015, 84: 551–575. DOI: 10.1146/annurev-biochem-060614-034142
|
| [6] |
Ahnert S E, Marsh J A, Hernández H, et al. Principles of assembly reveal a periodic table of protein complexes. Science, 2015, 350 (6266): aaa2245. DOI: 10.1126/science.aaa2245
|
| [7] |
Ellis R J. Protein misassembly. In: Csermely P, Vígh L, editors. Molecular Aspects of the Stress Response: Chaperones, Membranes and Networks. New York, NY, USA: Springer New York, 2007 : 1−13.
|
| [8] |
Tompa P, Rose G D. The Levinthal paradox of the interactome. Protein Science, 2011, 20 (12): 2074–2079. DOI: 10.1002/pro.747
|
| [9] |
Zhu J, Avakyan N, Kakkis A, et al. Protein assembly by design. Chemical Reviews, 2021, 121 (22): 13701–13796. DOI: 10.1021/acs.chemrev.1c00308
|
| [10] |
Bergendahl L T, Gerasimavicius L, Miles J, et al. The role of protein complexes in human genetic disease. Protein Science, 2019, 28 (8): 1400–1411. DOI: 10.1002/pro.3667
|
| [11] |
Kennedy K A, Gachelet E G, Traxler B. Evidence for multiple pathways in the assembly of the Escherichia coli maltose transport complex. Journal of Biological Chemistry, 2004, 279 (32): 33290–33297. DOI: 10.1074/jbc.M403796200
|
| [12] |
Britt H M, Cragnolini T, Thalassinos K. Integration of mass spectrometry data for structural biology. Chemical Reviews, 2022, 122 (8): 7952–7986. DOI: 10.1021/acs.chemrev.1c00356
|
| [13] |
Zhang S, Zou S, Yin D, et al. USP14-regulated allostery of the human proteasome by time-resolved cryo-EM. Nature, 2022, 605: 567–574. DOI: 10.1038/s41586-022-04671-8
|
| [14] |
Bansal P K, Abdulle R, Kitagawa K. Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Molecular and Cellular Biology, 2004, 24 (18): 8069–8079. DOI: 10.1128/MCB.24.18.8069-8079.2004
|
| [15] |
Ishii K, Zhou M, Uchiyama S. Native mass spectrometry for understanding dynamic protein complex. Biochimica et Biophysica Acta(BBA)-General Subjects, 2018, 1862 (2): 275–286. DOI: 10.1016/j.bbagen.2017.09.019
|
| [16] |
Hernández H, Robinson C V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature Protocols, 2007, 2 (3): 715–726. DOI: 10.1038/nprot.2007.73
|
| [17] |
Chorev D S, Baker L A, Wu D, et al. Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science, 2018, 362 (6416): 829–834. DOI: 10.1126/science.aau0976
|
| [18] |
Levy E D, Erba E B, Robinson C V, et al. Assembly reflects evolution of protein complexes. Nature, 2008, 453: 1262–1265. DOI: 10.1038/nature06942
|
| [19] |
Marsh J A, Hernández H, Hall Z, et al. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell, 2013, 153 (2): 461–470. DOI: 10.1016/j.cell.2013.02.044
|
| [20] |
Levy E D, Pereira-Leal J B, Chothia C, et al. 3D complex: a structural classification of protein complexes. PLoS Computational Biology, 2006, 2 (11): e155. DOI: 10.1371/journal.pcbi.0020155
|
| [21] |
Peterson L X, Togawa Y, Esquivel-Rodriguez J, et al. Modeling the assembly order of multimeric heteroprotein complexes. PLoS Computational Biology, 2018, 14 (1): e1005937. DOI: 10.1371/journal.pcbi.1005937
|
| [22] |
Esquivel- Rodríguez J, Yang Y D, Kihara D. Multi-LZerD: multiple protein docking for asymmetric complexes. Proteins, 2012, 80 (7): 1818–1833. DOI: 10.1002/prot.24079
|
| [23] |
Jones G. Genetic and evolutionary algorithms. In: Schleyer P von R, Allinger N L, Clark T, et al, editors. Encyclopedia of Computational Chemistry. Chichester, UK: John Wiley & Sons, Ltd. 1998 .
|
| [24] |
Kurisaki I, Tanaka S. Computational prediction of heteromeric protein complex disassembly order using hybrid Monte Carlo/molecular dynamics simulation. Physical Chemistry Chemical Physics, 2022, 24 (17): 10575–10587. DOI: 10.1039/D2CP00267A
|
| [25] |
Takada S. Coarse-grained molecular simulations of large biomolecules. Current Opinion in Structural Biology, 2012, 22 (2): 130–137. DOI: 10.1016/j.sbi.2012.01.010
|
| [26] |
Pak A J, Voth G A. Advances in coarse-grained modeling of macromolecular complexes. Current Opinion in Structural Biology, 2018, 52: 119–126. DOI: 10.1016/j.sbi.2018.11.005
|
| [27] |
Takada S, Kanada R, Tan C, et al. Modeling structural dynamics of biomolecular complexes by coarse-grained molecular simulations. Accounts of Chemical Research, 2015, 48 (12): 3026–3035. DOI: 10.1021/acs.accounts.5b00338
|
| [28] |
Li W F, Wang W, Takada S. Energy landscape views for interplays among folding, binding, and allostery of calmodulin domains. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111 (29): 10550–10555. DOI: 10.1073/pnas.1402768111
|
| [29] |
Kenzaki H, Koga N, Hori N, et al. CafeMol: A coarse-grained biomolecular simulator for simulating proteins at work. Journal of Chemical Theory and Computation, 2011, 7 (6): 1979–1989. DOI: 10.1021/ct2001045
|
| [30] |
Nikam R, Kulandaisamy A, Harini K, et al. ProThermDB: thermodynamic database for proteins and mutants revisited after 15 years. Nucleic Acids Research, 2020, 49 (D1): D420–D424. DOI: 10.1093/nar/gkaa1035
|
| [31] |
Gaudet R, Savage J R, McLaughlin J N, et al. A molecular mechanism for the phosphorylation-dependent regulation of heterotrimeric G proteins by phosducin. Molecular Cell, 1999, 3 (5): 649–660. DOI: 10.1016/S1097-2765(00)80358-5
|
| [32] |
Loew A, Ho Y K, Blundell T, et al. Phosducin induces a structural change in transducin βγ. Structure, 1998, 6 (8): 1007–1019. DOI: 10.1016/S0969-2126(98)00102-6
|
| [33] |
Blüml K, Schnepp W, Schröder S, et al. A small region in phosducin inhibits G-protein βγ-subunit function. The EMBO Journal, 1997, 16 (16): 4908–4915. DOI: 10.1093/emboj/16.16.4908
|
| [34] |
Danner S, Lohse M J. Phosducin is a ubiquitous G-protein regulator. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93 (19): 10145–10150. DOI: 10.1073/pnas.93.19.10145
|
| [35] |
Zheng S, Qin F, Yin J, et al. Role and mechanism of actin-related protein 2/3 complex signaling in cancer invasion and metastasis: A review. Medicine, 2023, 102 (14): e33158. DOI: 10.1097/MD.0000000000033158
|
| [36] |
Robinson R C, Turbedsky K, Kaiser D A, et al. Crystal structure of Arp2/3 complex. Science, 2001, 294 (5547): 1679–1684. DOI: 10.1126/science.1066333
|
| [37] |
Webb B, Sali A. Comparative protein structure modeling using MODELLER. Current Protocols in Bioinformatics, 2016, 54: 5.6. 1–5.6. 37. DOI: 10.1002/cpbi.3
|
| [38] |
Pollard T D, Beltzner C C. Structure and function of the Arp2/3 complex. Current Opinion in Structural Biology, 2002, 12 (6): 768–774. DOI: 10.1016/S0959-440X(02)00396-2
|
| [39] |
Fäßler F, Dimchev G, Hodirnau V V, et al. Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction. Nature Communications, 2020, 11: 6437. DOI: 10.1038/s41467-020-20286-x
|
| [40] |
Gournier H, Goley E D, Niederstrasser H, et al. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Molecular Cell, 2001, 8 (5): 1041–1052. DOI: 10.1016/S1097-2765(01)00393-8
|
| Subunit | Gtβ | Gtγ | phosducin | Sum |
| Gtβ c(339) | a327 | 308 | b635 | |
| Gtγ (65) | 3 | 329 | ||
| phosducin (188) | 311 | |||
| aNative contacts between two subunits; bthe total number of intersubunit native contacts of the subunit; cthe number in parentheses is the number of residues of the subunit. | ||||
| Subunit | ARPC4 | ARPC2 | Arp3 | Arp2 | ARPC1 | ARPC5 | ARPC3 | Sum |
| ARPC4 c(167) | a244 | 72 | 102 | 188 | 107 | 0 | b713 | |
| ARPC2 (282) | 213 | 0 | 2 | 0 | 0 | 459 | ||
| Arp3 (414) | 79 | 0 | 0 | 88 | 452 | |||
| Arp2 (378) | 37 | 98 | 0 | 316 | ||||
| ARPC1 (368) | 80 | 0 | 307 | |||||
| ARPC5 (143) | 0 | 285 | ||||||
| ARPC3 (173) | 88 | |||||||
| aNative contacts between two subunits; bthe total number of intersubunit native contacts of the subunit; cthe number in parentheses is the number of residues of the subunit. | ||||||||